Introduction
The oral cavity is a microorganism rich ecosystem; various conditions can disturb the homeostatic balance by creating ecological stress in the oral cavity, facilitating the emergence and growth of perio pathogenic and cariogenic bacteria. 1 During the orthodontic treatment of various malocclusions using fixed orthodontic appliances may influence change in the oral environment. 2 The changes in the oral microflora can potentially lead to increased risk of gingivitis, periodontal disease and demineralization of teeth and white spot lesions. 3 Subgingival periodontal biofilm/plaque is accumulated with as Aggregatibacter actinomycetemcomitans (AA), Porphyromonas gingivalis(PG), Prevotella intermedia(PI), Tannerella forsythia(TF), and Treponema denticola (TD) causing progressive periodontitis. 4
Two types of brackets are commonly used in orthodontics practice namely PEA and self-ligating brackets. The subgingival microflora in deepened periodontal pockets is dominated by Gram- negative anaerobic rods and spirochetes. Aggregatibacter actinomycetemcomitans [AA], Porphyromonas gingivalis [PG], are active markers for periodontitis in adults and these species were linked to succession of the disease. As technology progress, more sensitive techniques based on DNA-amplification such as the Polymerase Chain Reaction, have been developed. 5, 6, 7
Numerous studies show sustainable reproducibility of a commercial multiplex PCR-based test for the observation and semi-quantification of subgingival periodontal pathogenic species. 8, 9 Orthodontic treatment can suppress pathologic tooth migration, control bacterial plaque, and establish a good occlusion to promote the restoration of the periodontal tissues.10, 11, 12 Therefore, The aim of this study was to evaluate and compare the levels of Aggregatebacter actinomycetemcomitans [AA] and Porphyromonas gingivalis [PG] in the gingival crevicular fluids of the patients undergoing orthodontic treatment with PEA(3M unitek) and self-ligating brackets(Damon Q; ORMCO).
Materials and Methods
The sample was selected from the patients who had reported to the Department of Orthodontics AlBadar Rural Dental College and Hospital, Kalaburagi, Karnataka. The sample size has been estimated using the GPower software v. 3.1.9.4(Franz Faul, Universität Kiel, Germany) considering the effect size to be measured(f) at 24% power of the study at 80% and the alpha error at 5%, the sample size needed was 30. Each study group comprise of 15 patients [15 samples x 2 groups = 30 samples].The inclusion and exclusion criteria are:-
Inclusion criteria
Individuals with healthy periodontium with general probing depth of <3mm.
Age group around 12 - 20 years.
Fixed appliance in both the arches.
Non smoker.
No oral habits reported.
PEA(MBT Prescription; 3M Unitek) and Passive Self-ligating Brackets (Damon Q; ORMCO) with 0.022x0.028 inch slot dimension.
Exclusion criteria
Any active carious lesions.
Topical fluoride application(except for a fluoridated dentifrice or antibacterial therapy within 6 months.
Any systematic disease.
The GCF samples were collected at baseline(Before the fixation of orthodontic appliance), 3 months and 6 months after initial orthodontic activation with the corresponding initial Cu NiTi wires from each group. Bonding of the brackets was done by direct technique with light-cured adhesive(Transbond XT, 3M Unitek), excess composite was removed from the tooth surface. The archwire were fully engaged in each bracket after bonding.
GCF sample was taken from identical crevicular locations of mandibular incisors from the same site at 3 different time-points i.e, baseline, 3 months 6 months from the mesial and distal surfaces of the anterior teeth of 30 (15 per group ie., 15 for Self-ligating Brackets(Damon Q; ORMCO) and 15 for PEA (MBT Prescription; 3M Unitek) patients. . The GCF was collected using an absorbent paper strip(perio paper, Pro-Flow; Interstate Drug Exchange, Amityville, NY) placed into the sulcus until some resistance is felt and left it for 30 seconds till the paper strips are completely filled (Figure 1). The filled strip was placed in the tube, 1.5-2 ml effendroff tube.
It was then placed in a 50‐55°C water bath or heating block for 1 hour. After 1 hour, 0.1ml DNA Salt Solution was added to the tube and mix by inverting the tube several times. Then Centrifuge(Figure 2) the tube for 5 minutes at 5,000 xg to pellet the cell debris.0.8ml Precipitation Solution were added , close the effendroff tube and, whilst watching, slowly invert the tube several times to mix. White DNA strands may appear(Figure 3).
Then the Polymerase chain Reaction(PCR) (Figure 4) was carried out in 0.2 ml PCR tubes in a thermal cycler. The 10 ml GCF bacterial DNA extract and controls was amplified with 0.5 mM(3F and 3R) primers. 200 mM of each dNTP (Promega),10 mM KCl PCR buffer, 2 mM MgCl2 and 1.0 U Taq polymerase(Bioline).Amplification conditions for both PCRs are as follows: 5 min at 94 uC to denature the DNA, followed by 40 cycles of denaturation at 94 uC for 1 min, primer annealing at 55 uC for 1 min and strand extension at 72 uC for 2 min on a thermal cycler. PCR products was separated on a 1.5 % agarose gel and DNA bands was visualized with ethidium bromide. The gel was stained(Figure 5) with 0.5 mg/ml ethidium bromide, viewed under UV transilluminator and images are captured on a gel documentation system.
Statistical Package for Social Sciences [SPSS] for Windows Version 22.0 Released 2013. Armonk, NY: IBM Corp., was used to perform statistical analyses. Independent Student t-Test / Mann Whitney Test was used to compare the mean adhesion of periodontal pathogens between 2 groups at different time intervals. Repeated Measures of ANOVA followed by Bonferroni’s Post hoc Test followed by Wilcoxon Signed Rank Post hoc test was used to compare the mean adhesion of periodontal pathogens between different time intervals in each study group. The level of significance [P-Value] was set at P<0.05.
Results
The evaluation of the colonization of PG and AA during the OTM using MBT and SLB was carried out to analyse their concentrations in GCF at baseline, 3 months and 6 months of orthodontic treatment. The data analysis including the colony forming units(CFUs) and DNA concentration in both MBT and SLB system was done which is tabulated from(Table 1, Table 2, Table 3, Table 4, Table 5, Table 6).
The descriptive analysis showed P. gingivalis bacterial count after placing the MBT Bracket at baseline was 1.53±0.32 for both the bracket system and at 3 months it was 2.58±0.20×102 CFU/ml and SLB was 2.09±0.4× 102 CFU/ml. At 6 Months was 29.53±3.02×103 CFU/ml and SLB was 2.11±0.17× 102 CFU/ml
The mean CFUs of P. gingivalis in MBT system showed a significant difference between time intervals and the difference was statistically highly significant at p<0.001. Multiple comparison of mean differences between time intervals revealed that the CFUs was significantly lesser in T0 time interval as compared to T1 and T2 and the mean differences were statistically highly significant at p<0.001. This was then followed next by T1 time interval which showed significantly lesser mean CFUs as compared to T2 Time interval and the mean difference was statistically highly significant at p<0.001(Table 7).
This infers that the mean CFUs of P. gingivalis in SLB system significantly increased with increase in time intervals from T0 to T2, with no difference between T1 and T2.compared to T1 and T2 and the mean differences were statistically highly significant at p<0.001.The mean CFUs of P. gingivalis in SLB system showed a significant difference between time intervals and the difference was statistically highly significant at p<0.001.
Multiple comparison of mean differences between time intervals revealed that the mean CFUs was significantly lesser in T0 time interval as compared to T1 and T2 and the mean differences were statistically significant at p=0.005 and p<0.001 respectively. However, the mean CFUs did not show significant difference between T1 and T2 time intervals(Table 8).
The DNA concentration level was highest for MBT group at T2 followed by T1. In MBT system, between time intervals, this difference was highly significantly(< 0.001) increased from T0 to T1 and significantly from T1 to T2 for PG and AA. In SLB system, the mean DNA concentration level value of PG significantly increased with increase in time intervals from T0 to T2, with no significant difference between T1 to T2.
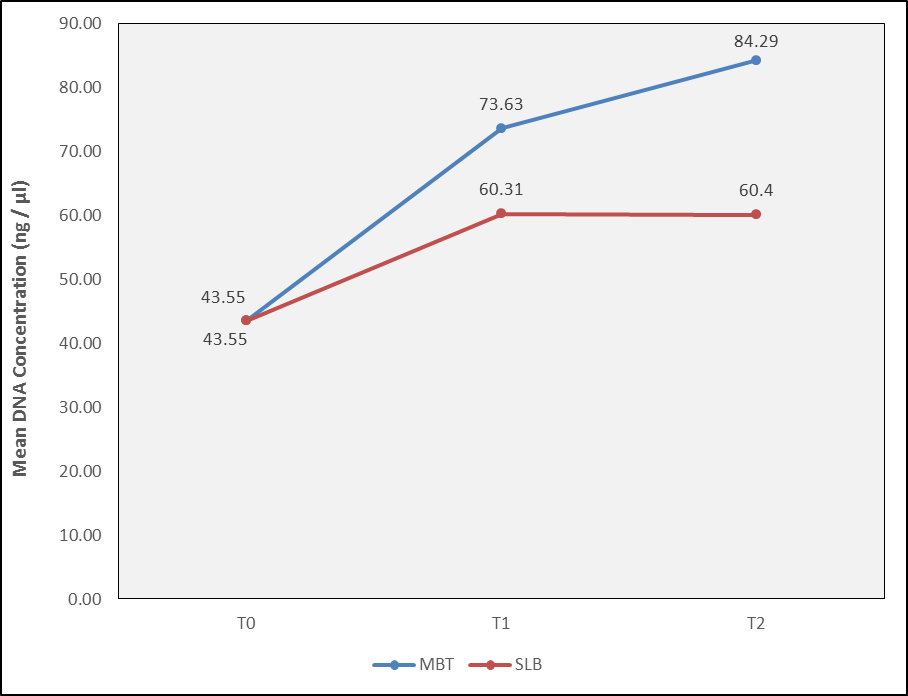
The mean DNA concentration level of AA remained the same from T0 to T1 to T2 (p value –0.27).The descriptive analysis of DNA Concentration(ng/µl) of P. gingivalis in MBT brackets at T0 was 43.55±9.06 ng/μl, after 3 months time interval 73.63±5.63 ng/μl and at 6 months was 84.29±8.62 ng/μl. In SLB brackets At T0 was 43.55±9.06and T1 was 60.31±11.86 ng/μl and At T2 was 60.4±4.8ng/μl (Figure 6).
The descriptive analysis DNA concentration(ng/µl) of A. actinomycetemcomitans in MBT Brackets at T0 was 43.76±7.68 ng/μl, At T1 time interval 62.41±12.05 ng/μl and at 6 months(T2) was 86.00±11ng/μl. In SLB brackets T0 was 43.76±7.68 ng/μl, 44.86±7.4 ng/μl at 3 months and 45.47±6.6 ng/μl at 6 months(Figure 7).
The mean DNA Concentration of A. actinomycetemcomitans in MBT system showed a significant difference between time intervals and the difference was statistically highly significant at p<0.001. This infers that the mean DNA Concentration of A. actinomycetemcomitans in MBT system significantly increased with increase in time intervals from T0 to T2.The mean DNA concentration of A. actinomycetemcomitans in SLB system showed no significant difference between time intervals [p=0.27].
Table 1
Comparisonof mean CFUs/ml(x 102) of P. gingivalis and A. actinomycetemcomitans between MBT and SLB at T0 time interval.
|
Organism |
Group |
N |
Mean |
SD |
Mean Diff |
p-value |
|
PG |
MBT |
15 |
1.53 |
0.32 |
0.00 |
1.00 |
|
SLB |
15 |
1.53 |
0.32 |
|||
|
AA |
MBT |
15 |
1.53 |
0.27 |
0.00 |
1.00 |
|
SLB |
15 |
1.53 |
0.27 |
Table 2
Comparison of mean DNA Concentration(ng/µl) of P. gingivalis and A. actinomycetemcomitans between MBT and SLB System at T0 time interval.
|
Organism |
Group |
N |
Mean |
SD |
Mean Diff |
p-value |
|
PG |
MBT |
15 |
43.552 |
9.055 |
0.00 |
1.00 |
|
SLB |
15 |
43.552 |
9.055 |
|||
|
AA |
MBT |
15 |
43.761 |
7.678 |
0.00 |
1.00 |
|
SLB |
15 |
43.761 |
7.678 |
Table 3
Comparison of mean CFUs/ml (x102) of P. gingivalis and A. actinomycetemcomitans between MBT and SLB System at T1 time interval.
|
Organism |
Group |
N |
Mean |
SD |
Mean Diff |
p-value |
|
PG |
MBT |
15 |
2.58 |
0.20 |
0.49 |
<0.001* |
|
SLB |
15 |
2.09 |
0.43 |
|||
|
AA |
MBT |
15 |
2.19 |
0.42 |
0.72 |
<0.001* |
|
SLB |
15 |
1.58 |
0.26 |
Table 4
Comparison of mean DNA Concentration(ng/µl) of P. gingivalis and A. actinomycetemcomitans between MBT and SLB System at T1 time interval.
|
Organism |
Group |
N |
Mean |
SD |
Mean Diff |
p-value |
|
PG |
MBT |
15 |
73.633 |
5.626 |
13.32 |
<0.001* |
|
SLB |
15 |
60.315 |
11.859 |
|||
|
AA |
MBT |
15 |
62.408 |
12.054 |
20.55 |
<0.001* |
|
SLB |
15 |
44.859 |
7.448 |
Table 5
Comparison of mean CFUs/ml(x102) of P. gingivalis and A. actinomycetemcomitans between MBT and SLB System at T2 time interval.
|
Organism |
Group |
N |
Mean |
SD |
Mean Diff |
p-value |
|
PG |
MBT |
15 |
29.53 |
3.02 |
27.43 |
<0.001* |
|
SLB |
15 |
2.11 |
0.17 |
|||
|
AA |
MBT |
15 |
30.13 |
3.98 |
28.54 |
<0.001* |
|
SLB |
15 |
1.59 |
0.23 |
Table 6
Comparison of mean DNA Concentration(ng/µl) of P. gingivalis and A. actinomycetemcomitans between MBT and SLB System at T2 time interval.
|
Organism |
Group |
N |
Mean |
SD |
Mean Diff |
p-value |
|
PG |
MBT |
15 |
84.288 |
8.621 |
24.16 |
<0.001* |
|
SLB |
15 |
61.124 |
4.880 |
|||
|
AA |
MBT |
15 |
86.001 |
11.358 |
40.53 |
<0.001* |
|
SLB |
15 |
45.474 |
6.603 |
Table 7
Comparison of mean CFUs/ml(x102) of P. gingivalis between time intervals in MBT and SLB System.
Table 8
Comparison of mean CFUs/ml(x102) of A. actinomycetemcomitans between time interval in MBT and SLB System.
Discussion
Fixed orthodontic appliances can act as a retentive site for plaque buildup and are known to cause decreases in plaque pH and increase in salivary S. mutans and gingival inflammation. 3 It is believed that the progression of periodontitis is caused by several bacterial species, such as Aggregatibacter actinomycetemcomitans(AA), Porphyromonas gingivalis (PG), Prevotella intermedia(PI), Tannerella forsythia(TF), and Treponema denticola (TD), that accumulate in the subgingival periodontal biofilm/plaque. 13, 14 Analysis of microflora in GCF becomes more and more important in diagnosis and therapy of periodontal diseases.15, 16, 17 Periodontopathogens like Aggregatibacter actinomycetemcomitans (AA) and Porphyromonas gingivalis(PG) are present in the subgingival plaque. 18, 19 Orthodontic treatment is predominantly performed in juvenile and is considered for the study. 20, 21 Porphyromonas gingivalis are the late colonizers. 22
The realtime PCR is used in the study as anaerobic culture has limitations: it is time consuming and laborious, and it has a relatively low level of sensitivity. 23 The periodontal pathogens show 6% difference in their 16S rRNA genes compared to anaerobic cultures. Dewhirst FE et al., 2010 24 estimated that about 50% of oral bacterial species are resistant to cultivation and as such, the use of DNA-based techniques, such as 16S rDNA microarray, real-time PCR and checkerboard DNA-DNA hybridisation, is capable of identifying a different microbial profile compared to culture-dependent techniques.
The brackets influence the bacterial adhesion and biofilm. This may be attributed to the size, material type, design, and relevant physiochemical surface properties of the brackets. 25 Surface wettability is another critical factor for bacterial adhesion to underlying materials, as a material with higher surface wettability attracts more bacteria to its surface than a material with lower surface wettability. 26At all-time points, the bacterial counts for both P. gingivalis and A. actinomycetemcomitans are higher in MBT brackets compared to SLB brackets.
In MBT brackets the bacterial count tends to increase substantially over the observation period, indicating potential microbial proliferation and biofilm formation. The mean CFUs of P. gingivalis and A. actinomycetemcomitans in MBT system showed statistically significant increase from time intervals from T0 to T2. These findings are in concordance with Van Gastel et al., 2009. 27 who stated that the presence of a ligature rather than a clip around Conventional brackets may hinder effective plaque removal when compared with SLB leading to increased bacterial adhesion.
Türkkahraman et al., 2005 28 stated bacteria show higher affinities for elastomeric materials and ligature ties than stainless steel which are in concordance with my study. Pellegrini et al., 2009, 29 who conducted a study using a split-mouth design to compare self-ligating brackets(SLB) and conventional brackets(CB) in terms of bacterial retention. Their results showed that teeth bonded with SLB attachments had fewer bacteria in plaque compared to teeth bonded with edgewise appliances using elastomeric ligation.
According to Yang and coworkers, 2017, 30 the design of SLB is able to reduce microbial colonization and promote oral health due to the configuration without ligature which is in concordance with my study.The meta-analysis written by Maizeray and coworkers, 2021 31 assessed the efficiency of CB, passive and active SLB. The authors concluded that no differences between the three types of brackets had been seen. In terms of periodontal indices, they found less BOP for passive SLB compared to CB after 4–5 weeks after bonding.
According to Boyd and Baumrind et al., 1992, 32 Tufekci et al., 2011 33 Plaque retention increases after placement of fixed appliances, which is associated with increased risk of decalcification and gingival and periodontal changes which was in accordance with my study. Mouna Benkhalifa et al., 2022 34 agreed that orthodontic treatment, regardless of the type of appliance, caused quantitative and qualitative changes in the oral microbiome leading to an increase in the counts of cariogenic bacteria and periodontal pathogens that are associated with dental caries and periodontal disease. However, there were significant variations between the different types of appliances depending on their plaque-retaining properties and removability which was according to our study.
Present study is not in accordance with study conducted by Bergamo et al., 2017 35 that self-ligating brackets were associated with a higher incidence of periodontopathogens than conventional brackets because the bracket design seems to influence the levels of bacterial species involved in periodontal disease.
According to Mummolo S et al., 2013 36 the plaque index(PI) increased over time in each group as well as salivary flow, mostly in subjects treated with self- ligating brackets, suggesting a difference between conventional and self-ligating brackets. Bacteria showed a different trend of colonization in the two treated groups, as for subjects treated with conventional brackets it showed the greater value at the early stage of treatment(T1), followed by a decrease at T2 which is not in concordance with current study.
The concentration of P. gingivalis DNA tends to increase over time for both the MBT and SLB brackets. Across all time points, the MBT brackets generally exhibit higher concentrations of P. gingivalis DNA compared to the SLB brackets, which is not in accordance with study done by Marzie et al., 2022 1 who stated that P. gingivalis concentration was lesser in conventional brackets compared with self ligtaing.
The mean DNA Concentration of A. actinomycetemcomitans in SLB system was significantly lesser as compared to MBT system and the mean difference was statistically highly significant at p<0.001
MBT brackets tend to harbor higher bacterial counts and DNA concentrations for both P. gingivalis and A. actinomycetemcomitans compared to SLB brackets. SLB brackets exhibit low bacterial counts and DNA concentrations, indicating potentially reduced microbial colonization and biofilm formation compared to MBT brackets. Further in-situ studies with bracket raw materials are needed to provide valuable information on the relationships between bracket materials and bacterial adhesion as well as between oral pathogens and oral hygiene indices in orthodontic patients.
Conclusion
MBT brackets tend to foster higher bacterial growth of P. gingivalis and A. actinomycetemcomitans compared to SLB brackets due to use of elastomeric or stainless steel ligatures to secure the archwire within bracket slot. MBT brackets consistently exhibit higher bacterial counts(CFUs) compared to SLB brackets across all timelines, indicating a potentially greater propensity for bacterial colonization and biofilm formation. P. gingivalis and A. actinomycetemcomitans exhibit increased DNA concentrations over time in GCF, with the MBT brackets generally showing higher DNA concentrations compared to that of SLB brackets.